

Simultaneous PET-MRI has gained attention over the past five years due to the complementary advantages that can provide in the translational field of molecular imaging technologies. This is a highly interdisciplinary topic that involves new instrumentation, data acquisition strategies, image processing and reconstruction algorithms, bimodal contrast agents, preclinical and clinical diagnostic and therapeutic applications.
King's College London has been in three different consortia funded by the EU. It is our belief that sharing data with the scientific community will contribute to faster evolution of our field, and following consultation from other groups, we have decided to make available simulated PET-MR data that will enable validation of new strategies for image quantification with PET-MR. Additional datasets will be included in the future. The acquisition and availability of the data has been reviewed and approved by the St Thomas' and Guy's Research Ethics Committee (REC Study Number: 01/11/12).
The 4D PET simulations were devised to assess the accuracy and the properties of reconstruction and associated corrections for a specific count level similar to that observed in patients (Tsoumpas et al 2011).
The current version of the datasets includes five numerical phantoms were produced from real dynamic MR acquisitions during normal breathing cycle following the procedures described by Buerger et al 2012 and Tsoumpas et al 2011. Appropriate SUV and attenuation properties were defined as described in the paper. These values are comparable to values derived from a typical patient FDG scans, but set such that different tissues have different uptake and pathological parameters were introduced representing potential cancerous lesions.
In addition to the 4D PET simulations, we also provide dynamic MR images that can be useful to estimate respiratory motion. The reconstructions were performed using STIR (Software for Tomographic Image Reconstruction, http://stir.sf.net) release 2.1 (Thielemans et al 2012). The data were reconstructed with OSEM using 23 subsets and 2 iterations smoothed with an isotropic Gaussian filter of 4 mm FWHM. More detailed description of the specific data generation can be found in Buerger et al 2011.
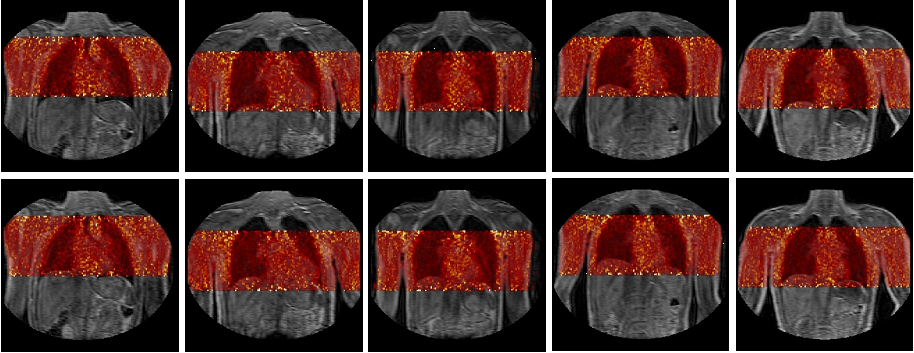
Fig. 1 PET-MR model-based emission maps of five volunteers at two different respiratory positions, end-inhale (top), end-exhale (bottom), Tsoumpas et al, 2011 © Institute of Physics, doi:10.1088/0031-9155/56/20/005
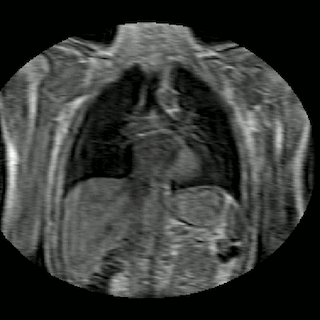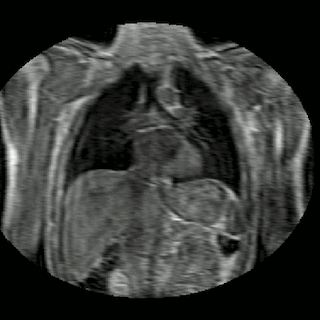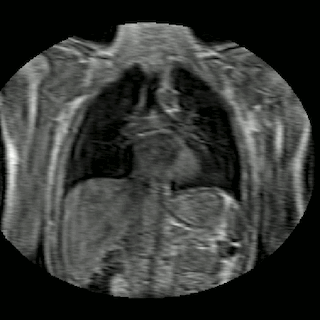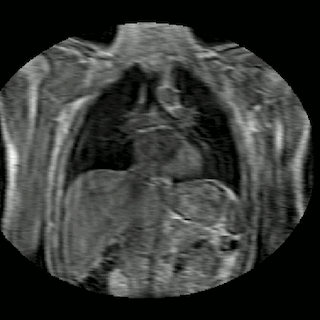
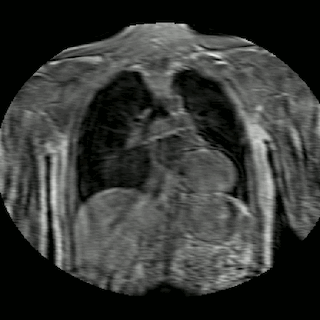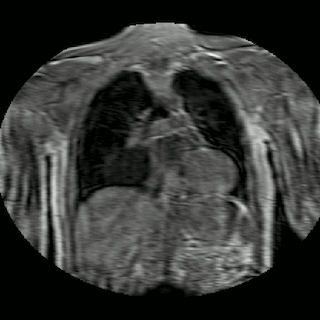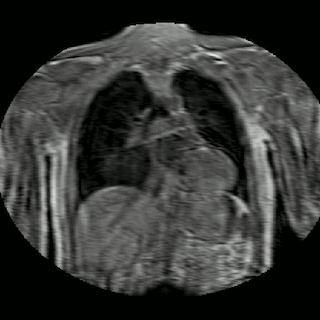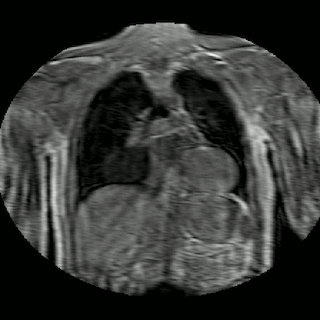
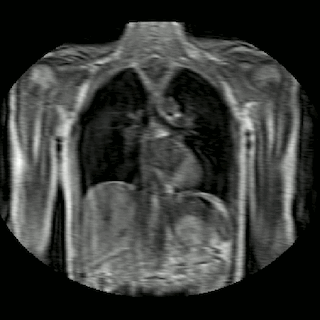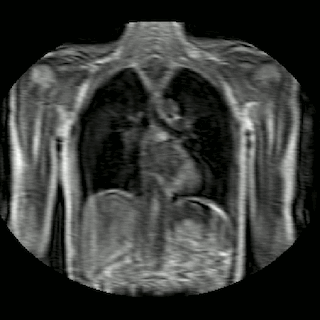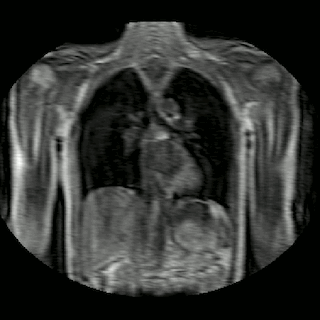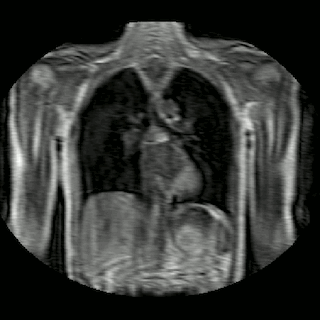
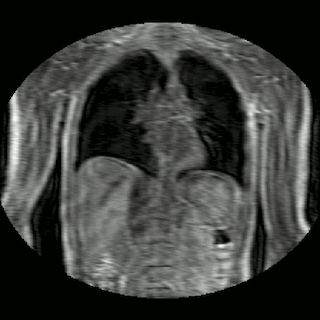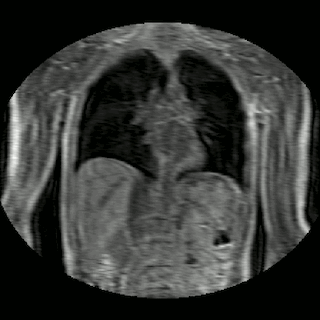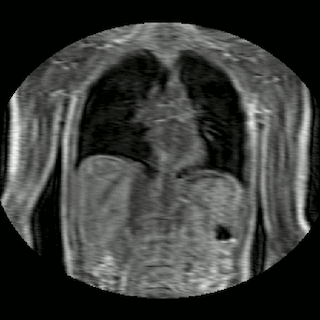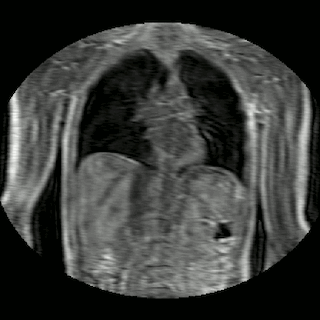
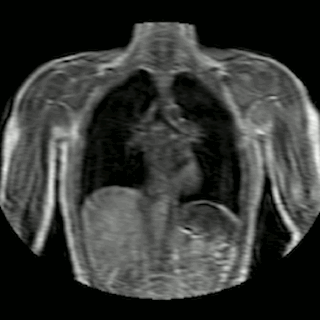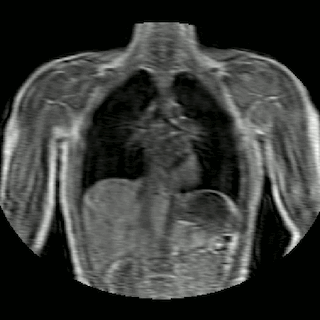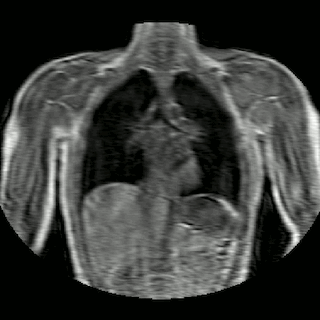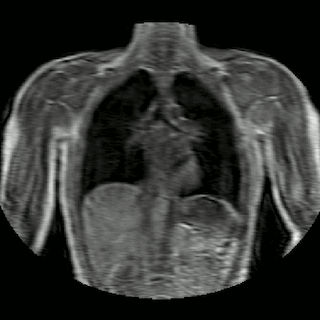
Fig 2. Animation of 4D MR data for five different volunteers.
The generation of the datasets is supported by HYPERimage (http://www.hybrid-pet-mr.eu) and SUBLIMA (http://www.sublima-pet-mr.eu), two independent EU funded projects under the seventh framework program (No: 201651, 241711) and the EU COST Action TD1007 (http://www.pet-mri.eu).